
Generative Topological Networks (GTNs)
Generative models have seen significant advancements in recent years, yet often remain challenging and costly to train and use. We introduce Generative Topological Networks (GTNs) -- a new class of generative models that addresses these shortcomings. GTNs are trained deterministically using a simple supervised learning approach grounded in topology theory. GTNs are fast to train, and require only a single forward pass in a standard feedforward neural network to generate samples. We demonstrate the strengths of GTNs on several datasets, including MNIST, CelebA and the Hands and Palm Images dataset. Additionally, the theory behind GTNs offers insights into how to train generative models for improved performance.
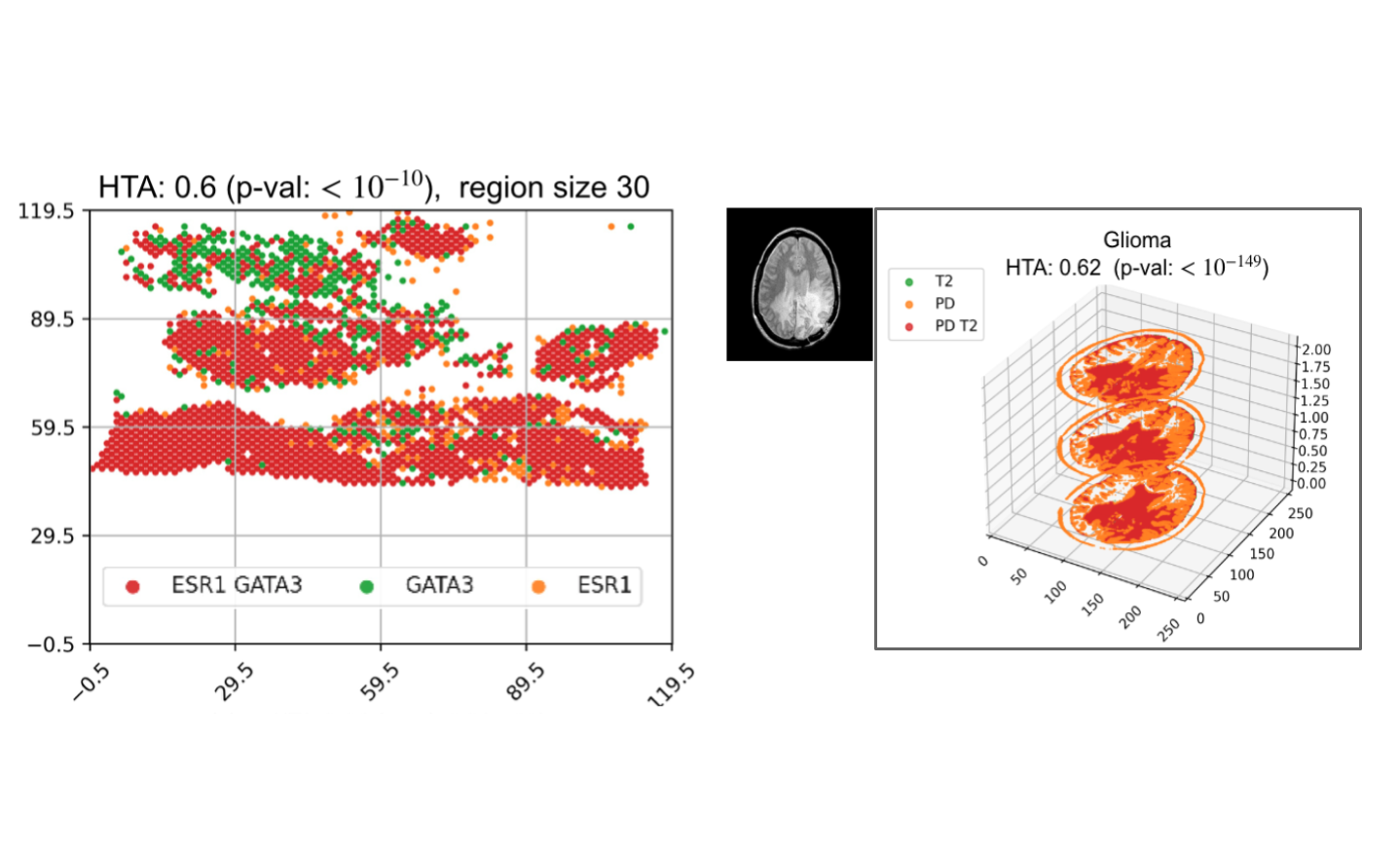
Assessing heterogeneity in spatial transcriptomics and imaging data using the HTA index
Emerging spatial transcriptomics data hold the potential to further our understanding of tumour heterogeneity and its clinical implications. Existing statistical tools are not sufficiently powerful to capture heterogeneity in the complex setting of spatial molecular biology. In this work, we developed the HeTerogeneity Average index (HTA), specifically designed to handle the multivariate nature of spatial transcriptomics. We prove that HTA has an approximately normal distribution, lending itself to efficient statistical assessment. Using HTA on two cancer spatial transcriptomics datasets we show that the spatial heterogeneity of the tumor is significantly linked to survival. We also apply HTA to 3D brain MRI data and show that HTA captures spatial differences between a normal ageing brain and different brain pathologies.
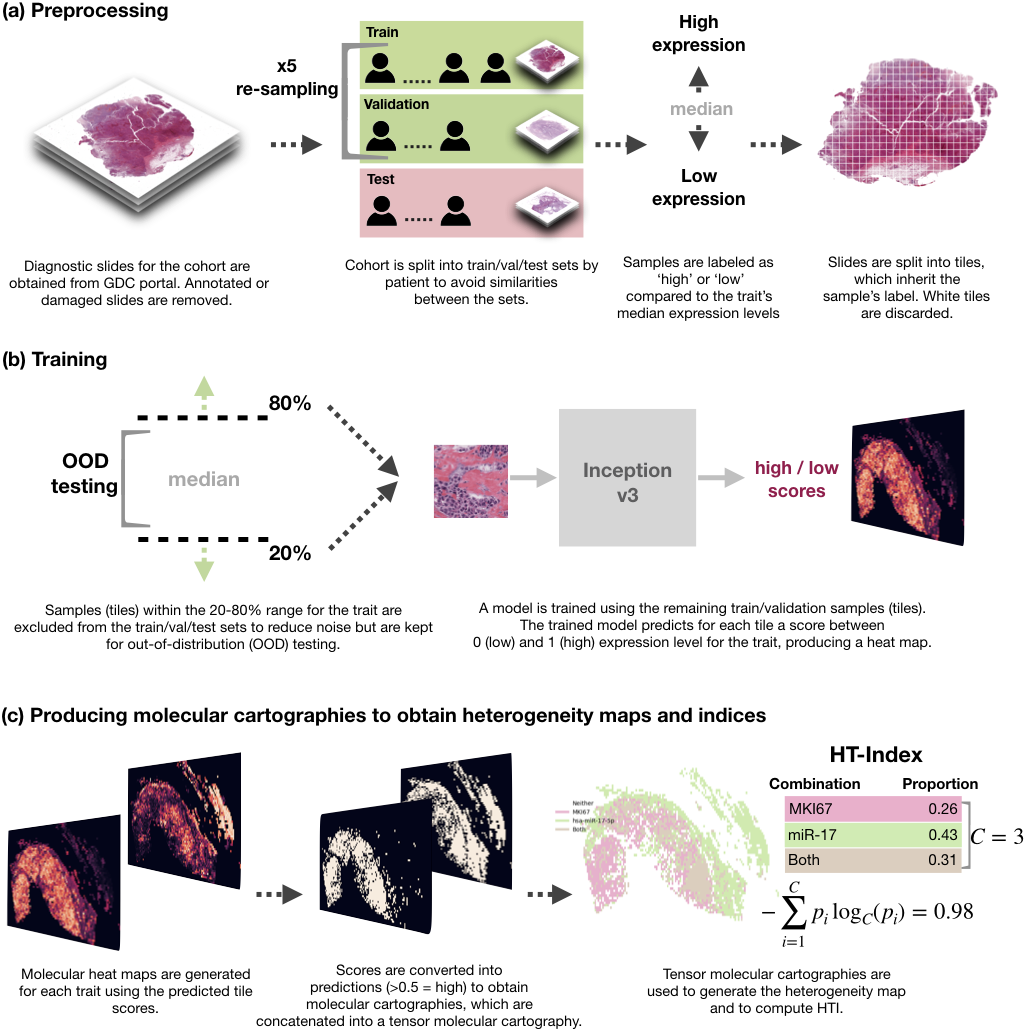
Spatial transcriptomics from pathology whole-slide images
In this work, we trained deep learning models to spatially resolve gene expression levels in pathology whole-slide images. Our models reach up to 0.95 AUC on held-out test sets from two cancer cohorts using a simple training pipeline and a small number of training samples. We also developed a method to spatially characterize tumor heterogeneity from the predicted gene expression levels. Applying our methods to breast and lung cancer slides, we discovered a significant statistical link between heterogeneity and survival. This project was supported by the European Union's Horizon 2020 and Google Cloud Platform.
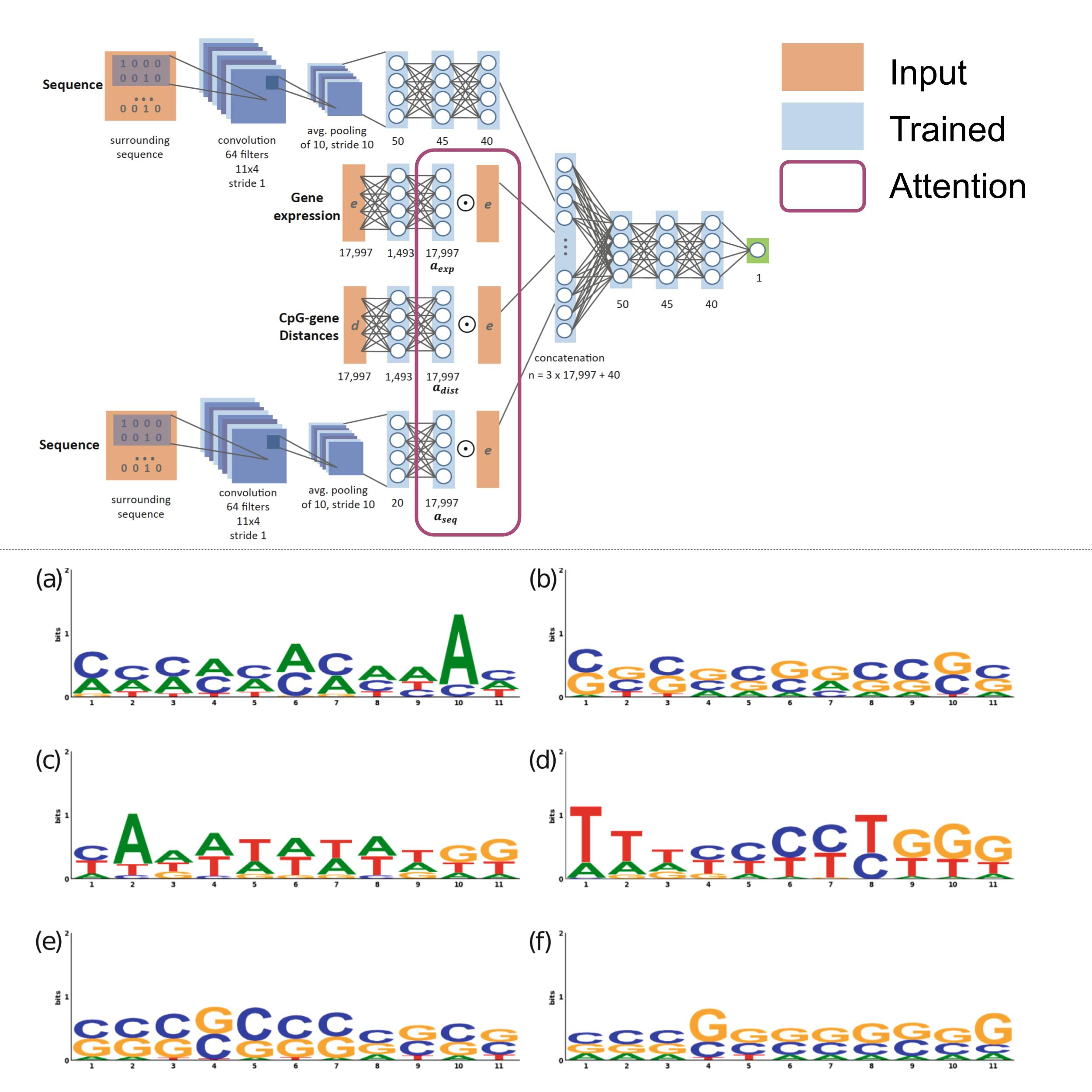
Predicting DNA methylation using deep learning with attention
DNA methylation has been extensively linked to alterations in gene expression, playing a key role in multiple diseases, especially cancer. In this work, we designed a deep learning network with an attention mechanism to predict methylation solely from sequence and gene expression. We also developed a framework to analyze the learned representations, which led to the discovery of novel links between methylation and gene expression. Our regression model obtains a Spearman correlation of 0.84 for thousands of CpG sites on two separate test sets and links several motifs and genes to methylation activity. The figure shows several DNA motifs identified by the network, including the TATA box and CpG islands, both known to play key roles in determining gene expression levels.
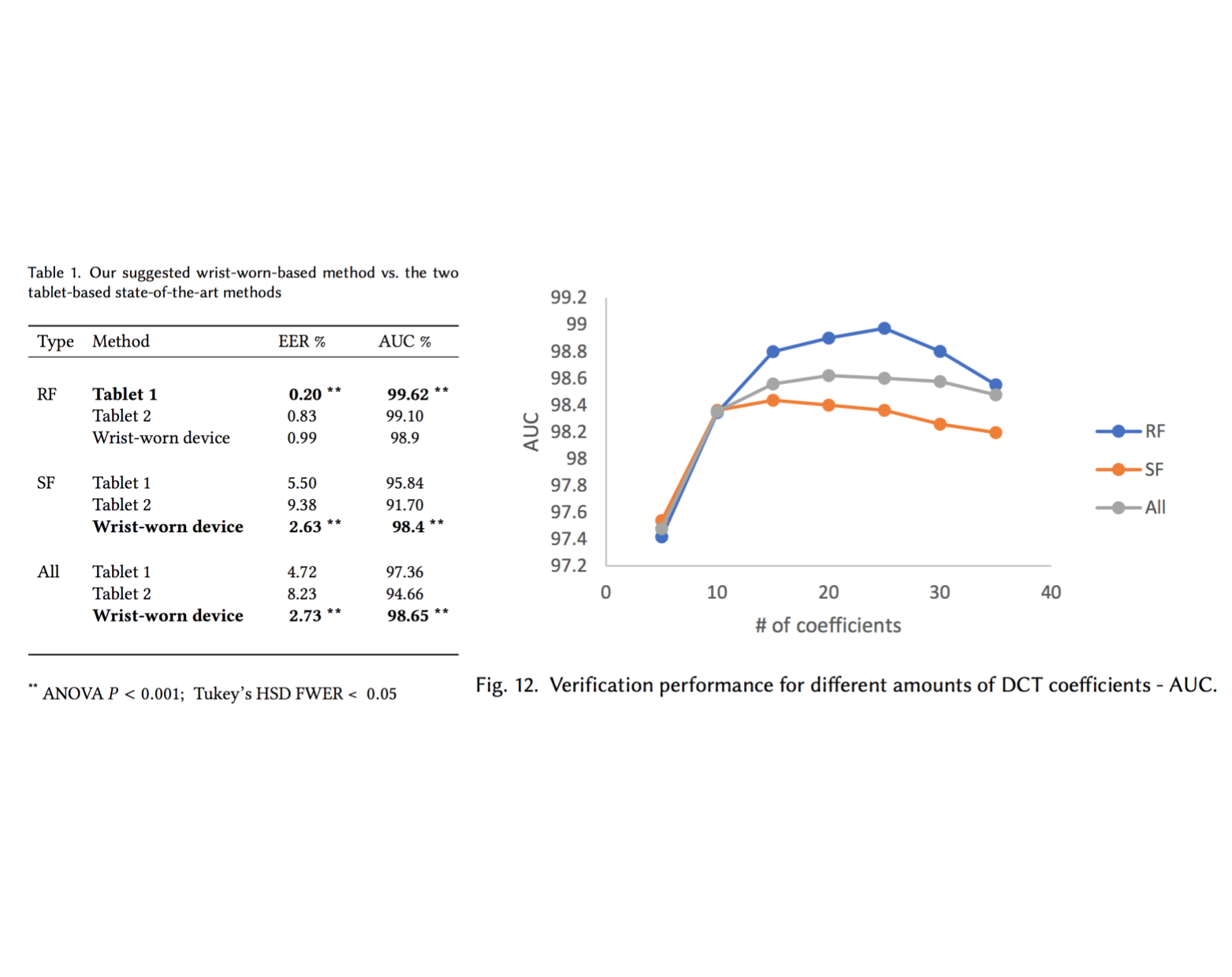
Handwritten signature verification using wrist-worn devices
In this work, we developed a machine learning model that identifies forged signatures based solely on motion signals captured from a smart-watch device (accelerometer, gyroscope etc.). To collect the data, we developed an android application that connects to a Microsoft Band 1, and used it to capture hundreds of signatures from 60 subjects. Our model significantly outperforms two state-of-the-art systems in skilled forgeries (SF), obtaining an EER of 2.63% and an AUC of 0.984. This paper won an outstanding student paper award (3rd place) in The 20th Israeli National Conference on Industrial Engineering.

